Rutaceae
Overview
| Family | Rutaceae |
| Genus | 160 (8 assembled genus) |
| Species | 1600 (24 assembled species) |
| SI type | Type-1 |
| SI genes | S-RNase (pistil specific expression); SLFs (pollen specific expression) |
Organism Image

Description
The Rutaceae is a family, commonly known as the rue or citrus family, of flowering plants, usually placed in the order Sapindales.
Species of the family generally have flowers that divide into four or five parts, usually with strong scents. They range in form and size from herbs to shrubs and large trees.
The most economically important genus in the family is Citrus, which includes the orange (C. × sinensis), lemon (C. × limon), grapefruit (C. × paradisi), and lime (various, mostly C. aurantifolia, the key lime). Boronia is a large Australian genus, some members of which are plants with highly fragrant flowers and are used in commercial oil production. Other large genera include Zanthoxylum, several species of which are cultivated for Sichuan pepper, Melicope, and Agathosma. About 160 genera are in the family Rutaceae.
SI type
The system with the broadest taxonomic distribution, which we term type-1 SI, is gametophytic and based on linked pistil S S-RNase and pollen S S-locus F-box (SLF)/S-haplotype-specific F-box (SFB). So far, type-1 SI has been found in four eudicot families: Solanaceae, Plantaginaceae, Rosaceae, and Rutaceae. After pollination, both S1- and S2-RNases can enter pollen tubes and be recognized by S1- or S3-SLFs, but only SCFS3-SLF complexes can ubiquitinate these S-RNases leading to their degradation by 26S proteasome, with the survived S1-RNase in S1 pollen tubes forming the S-RNase condensates (SRCs) resulting in self-pollen inhibition.
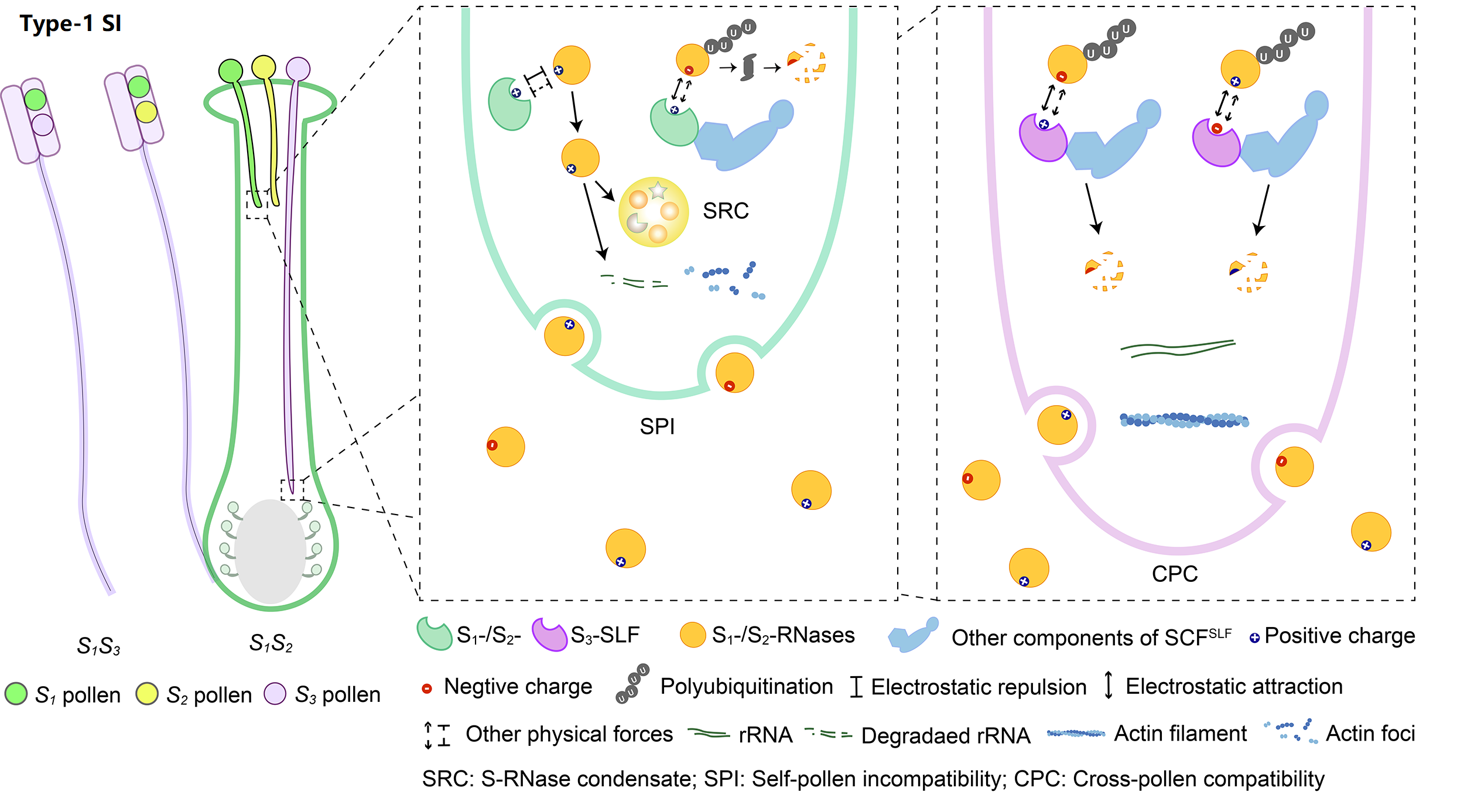
SI genes
Publication
Liang M, Cao Z, Zhu A, Liu Y, Tao M, Yang H, Xu Q Jr, Wang S, Liu J, Li Y, Chen C, Xie Z, Deng C, Ye J, Guo W, Xu Q, Xia R, Larkin RM, Deng X, Bosch M, Franklin-Tong VE, Chai L. Evolution of self-compatibility by a mutant Sm-RNase in citrus. Nat Plants. 2020 Feb;6(2):131-142. doi: 10.1038/s41477-020-0597-3.
de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin Heidelberg, Germany. https://link.springer.com/book/10.1007/978-3-662-04502-2https://link.springer.com/book/10.1007/978-3-662-04502-2
Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467-89. doi: 10.1146/annurev.arplant.56.032604.144249.
Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annu Rev Plant Biol. 2009;60:21-42. doi: 10.1146/annurev.arplant.043008.092108.
Iwano M, Takayama S. Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol. 2012 Feb;15(1):78-83. doi: 10.1016/j.pbi.2011.09.003.
Fujii S, Kubo K, Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat Plants. 2016 Sep 6;2(9):16130. doi: 10.1038/nplants.2016.130.
Zhao H, Zhang Y, Zhang H, Song Y, Zhao F, Zhang Y, Zhu S, Zhang H, Zhou Z, Guo H, Li M, Li J, Gao Q, Han Q, Huang H, Copsey L, Li Q, Chen H, Coen E, Zhang Y, Xue Y. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022 Jan 20;34(1):579-596. doi: 10.1093/plcell/koab266.